Structure Based Rational Design
Structure based rational design involves using high quality 3D computer models of the interaction of a small organic molecule (ligand) with a large protein (enzyme or receptor) to produce either a new ligand or a new protein with altered properties. An important application of ligand design is Structure Based Drug Design (SBDD) in which an existing small molecule is logically modified, usually in an iterative fashion, into a drug candidate. In a key application, SBDD can be used to understand small molecule SAR; understanding SAR is the key to advancing SAR. An important application of protein design is Structure Based Enzyme Engineering (SBEE) in which and existing enzyme is logically modified, usually in an iterative fashion, into a new catalyst that is not found in nature.
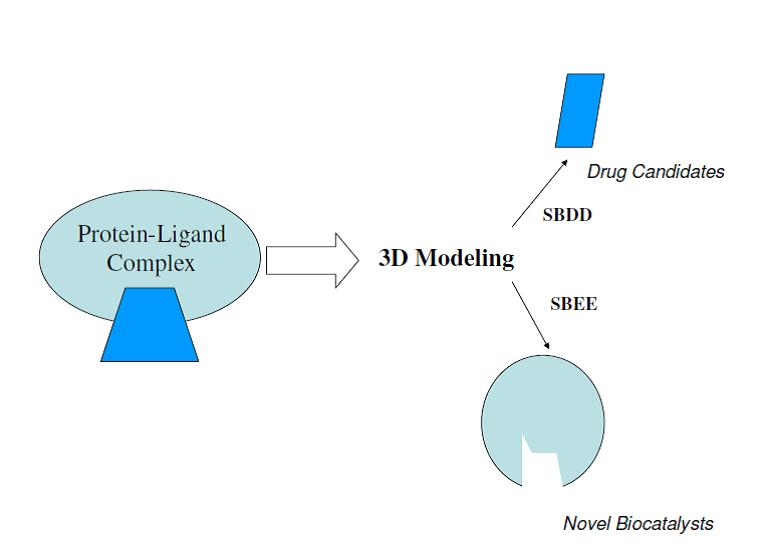
The key to both of these processes is the development of high quality 3D computational models for the protein-ligand complex. A high quality model is typically derived from an experimental (usually x-ray crystallography) structure of a related protein-ligand complex. For a high quality 3D model, in addition to structure it is crucial that function (how does the protein work and what does the ligand do) also be taken into account.
In quality SBDD programs several protein-ligand structures can be utilized in an iterative manner, both as a check of computational models and the refinement of these models. While having multiple experimental structures is useful in SBDD, it is not essential. For SBEE applications, while there is not an urgent need for multiple structures, more structural information will lead to better models.