Effectiveness of Protein Structure Based Design (SBDD) in Drug Discovery:
A Style and Approach that has proven to be
Very Successful
An informal literature mini review:
Robert E Babine
Rebexsess Discovery Chemistry
published on www.rebexsess.com, June 2011
Over the last 25 years or so protein structure has become an increasing important component of small molecules drug discovery programs. In many cases structures of protein-ligand complexes have been used to rationalize results after the fact (usually for publications) often because crystallography lags behind medicinal chemistry. However, protein crystallography can be effectively incorporated early in a discovery program.
(1) When protein crystallography is integrated early in the discovery phase it can have an enormous beneficial impact on a program.
(2) 3D protein-ligand complex structures allow for efficient optimization of small molecules, both with respect to protein-ligand optimization and equally important, approaches to effectively optimize drug like properties. Thus, more experts and professionals recognized its value through out pharmaceutical and biotech industry.
(3) In order for an integrated program to be a powerful tool for advancing small molecules into serious candidates for clinical studies, effective communication between crystallography, computational chemistry/molecular modeling and medicinal chemistry must be seamless.
A couple of successful case studies will now be reviewed.
Telaprevir (Incivek) is an oral protease inhibitor FDA approved for the treatment of
hepatitis C virus infection. It was
invented in a collaborative research program between Vertex and Eli Lilly. In this discovery program x-ray
crystallography (Vertex) was integrated into the program from the
beginning. Hepatitis C NS3
protease is a serine protease that is in a different structural class than the
trypsin class of proteases. This viral
protease has also evolved a Ser-His-Asp catalytic triad; however, biochemical
and structural data obtained in the early stages of the program indicated that
this protease behaved in a manner distinct from the trypsin class.
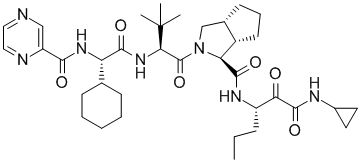
Telaprevir (Incivek)
In a significant effort, the Vertex
group, headed by Robert B. Perni, determined that keto-amides were effective inhibitors of this enzyme.
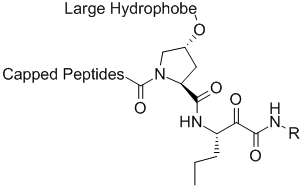
Vertex initial lead compound series
The Lilly group was able to use this vast amount of ligand SAR and protein structural information to interpret the SAR data in the context of protein-ligand interactions. The computational chemistry/molecular modeling support was able to facilitate effective communicate between crystallography and medicinal chemistry across different sites. (This situation is not unlike an outsourced program that might occur under the present environment). In one aspect, initiated by Mark J. Tebbe, the large and hydrophobic capped P2 groups of the Vertex initial lead series was targeted for simplification. An observation in crystallography that the histidine of the catalytic triad becomes less ordered upon binding keto-amides made this an especially attractive idea. Upon iterative discussions between medicinal/synthetic chemistry and computational chemistry/molecular modeling the 5,5-bicyclic pyrrolidine was designed. This bicyclic piece retained activity while reducing molecular weight and the number of rotatable bonds. This new novel P2 lead series was further optimized into the orally bioavailable clinical candidate telapravir.
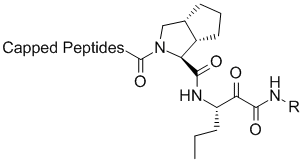
Lilly novel P2 lead series
The Vertex group developed telapravir through clinical trails and through FDA approval. The discovery program was supplemented from the beginning by Structure Based Design and it was a very useful tool throughout the discovery phase. The Schering group independently invented a related compound boceprevir (Victrelis, Merck), in a structure based discovery program. This compound was also approved by the FDA. Over the years, the 3D computational/modeling technique has been more and more recognized and accepted throughout the pharmaceutical and biotech industry as an effective and powerful drug discovery tool.
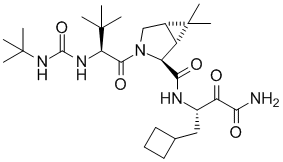
Boceprevir (Victrelis, Merck)
A decade before the discovery of Hepatitis C NS3 protease inhibitors (in several groups in addition to Vertex) another viral protease, HIV-1 protease, was the subject of an intense drug discovery effort that utilized Protein Structure Based Design (SBDD) as a useful drug discovery tool. One example is now discussed briefly.
In principle, SBDD allows one to go from the Discovery of new drugs (creation of new ligands by either following small molecule SAR of a series to serendipity) to the Invention of new drugs (the rational design of new entities based upon detailed knowledge of how small molecules interact with their protein target; serendipity can still come in handy). In practice, pure de novo design has proven extremely difficult; however, coupled with knowledge of substrates and/or known ligands, design from first principles can become practical.
HIV-1 protease is a C2 symmetric aspartic protease whose first known inhibitor was the natural product pepstatin. A design approach that incorporated the hydroxyl group from pepstatin into the center of a symmetric capped peptidomimetic led to novel inhibitor design. Thus, the thought process went from a symmetric protein to a symmetric inhibitor class via a bi-directional synthesis. This concept was executed to produce potent enzyme inhibitors quickly with a very small chemistry group. (Exploratory medicinal chemistry group at Lederle Laboratories initiated by Martin F. Semmelhack)
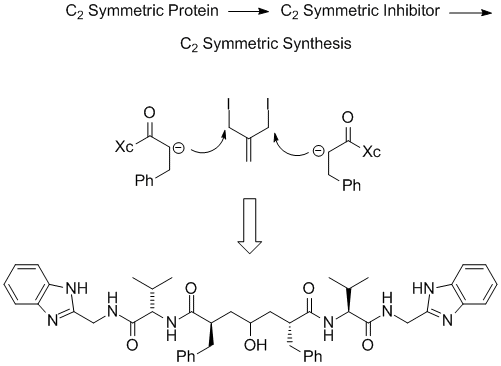
These symmetric inhibitors had poor physical and solubility properties and were not suitable for development. A medicinal chemistry group at Lederle, headed by Michael P. Trova and Allan Wissner, in iterative discussions with molecular modeling came up with the idea of combining a piece of the potent anti-viral Roche compound with the symmetric Lederle enzyme inhibitor. At that time, while the coordinates for neither compound were available, good models for both ligands were generated in house. Using molecular modeling, it became apparent how to combine these different compounds together. Execution of synthesis and extensive biological testing produced a novel hydroxylaminepentanamide inhibitor that was a potent antiviral and was orally bioavailble in rodents. This work demonstrates how knowledge of the structure of protein ligand complexes can assist in rational scaffold hopping between different ligand classes.
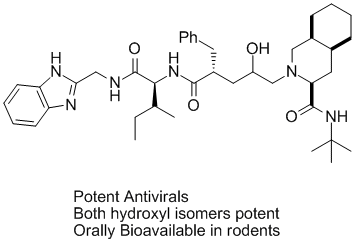
Hydroxylaminepentanamide inhibitors were independently developed by Merck and one member of this class (Crixivan) was FDA approved for the treatment of HIV-1 infection. Due to an uncertain patent situation the Lederle compounds were not developed further.
The above examples show a style and approach to drug discovery that has proven to be very successful.
- Introduction of protein structure early in a discovery program.
- Specifically, information of protein structure analyzed with appropriate computational tools, effectively communicated, allows a deep understanding of SAR that leads to effective advancement of that SAR.
- Close communication between protein structure determination and medicinal chemistry facilitated by computational chemistry/molecular modeling can be effective in the advancement of small molecules into drug candidates.
References and Notes:
Hepatitis C
“Discovery and development of VX-950, a novel, covalent, and reversible inhibitor of hepatitis C virus NS3.4A serine protease C. Lin, A.D. Kwong AD and R.B. Perni Infect Disord Drug Targets. 2006, 6, 3-16.
"Peptidomimetic Protease Inhibitors"US Patent 7,820,671 Babine; Robert Edward, Chen; Shu
Hui, Collado; Ivan, Garcia-Paredes; Cristina, Glass; John Irvin, Guo;
Deqi, Jin; Ling ,
Lamar; Jason Eric, Parker, III; Raymond Samuel, Snyder; Nancy June, Sun; Xicheng David, Tebbe; Mark Joseph, Victor; Frantz , Wang; Q. May, Yip; Yvonne Yee Mai, Perni; Robert B., Farmer; Luc
(Composition of matter patent for telapravir)
The FDA has also approved boceprevir (Victrelis, Merck) for hepatitis C virus (HCV) infection.This compound has a similar structure and the discovery program had an active crystallography group:
see: “Discovery of (1R,5S)-N-[3-Amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]- 3-[2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (SCH 503034), a Selective, Potent, Orally Bioavailable Hepatitis C Virus NS3 Protease Inhibitor: A Potential Therapeutic Agent for the Treatment of Hepatitis C InfectionSrikanth Venkatraman, Stéphane L. Bogen, Ashok Arasappan, Frank Bennett, Kevin Chen, Edwin Jao, Yi-Tsung Liu, Raymond Lovey, Siska Hendrata, Yuhua Huang, Weidong Pan, Tejal Parekh, Patrick Pinto, Veljko Popov, Russel Pike, Sumei Ruan, Bama Santhanam, Bancha Vibulbhan, Wanli Wu, Weiying Yang, Jianshe Kong, Xiang Liang, Jesse Wong, Rong Liu, Nancy Butkiewicz, Robert Chase, Andrea Hart, Sony Agrawal, Paul Ingravallo, John Pichardo, Rong Kong, Bahige Baroudy, Bruce Malcolm, Zhuyan Guo, Andrew Prongay, Vincent Madison, Lisa Broske, Xiaoming Cui, Kuo-Chi Cheng, Yunsheng Hsieh, Jean-Marc Brisson, Danial Prelusky, Walter Korfmacher, Ronald White, Susan Bogdanowich-Knipp, Anastasia Pavlovsky, Prudence Bradley, Anil K. Saksena, Ashit Ganguly, John Piwinski, Viyyoor Girijavallabhan, and F. George Njoroge J. Med. Chem., 2006, 49, 6074 086
A Hepatitis C protease review:
"A review of HCV protease inhibitors" Chen, K. X.; Njoroge, F. G. Curr. Opin. Invest. Drugs 2009, 10, 821 837
Lederle HIV Protease:
"Synthesis and Biological Evaluation of a Series of HIV-1
Protease Inhibitors" Michael P.Trova, Robert E.
Babine,Randal A. Byrn, Wellington T. Casscles, Richard C. Hastings, Grace C. Hsu, Michael R.
Jirousek, Bernard D. Johnson, Suresh S. Kerwar,
Steven R. Schow, Allan Wissner,
Nan Zhang and Michael M. Wick Bioorg. Med.
Chem. Lett. 1993, 3, 1595-1600
"Structure Activity Studies on Pseudo-symmetrical HIV-1
Protease Inhibitors" Robert E. Babine, Nan Zhang, Steven R. Schow, Suresh S. Kerwar, Parimal R. Desai, Randal A. Byrn
and Richard C. Hastings Bioorg. Med. Chem.
Lett.1993, 3, 1590-5
"Retroviral Protease Inhibitors"
The Merck path to Crixivan
followed a similar (independent) path:
"X-ray crystal structure of the HIV protease complex with L-700,417, an inhibitor with pseudo C2 symmetry" Roger Bone, Joseph P. Vacca, Paul S. Anderson, M. Katharine Holloway J. Am. Chem. Soc., 1991, 113, 9382-384
"L-735,524: The Design of a Potent and Orally Bioavailable HIV Protease Inhibitor" Bruce D. Dorsey, Rhonda B. Levin, Stacy L. McDaniel, Joseph P. Vacca, James P. Guare, Paul L. Darke, Joan A. Zugay, Emilio A. Emini, William A. Schleif J. Med. Chem., 1994, 37, 3443-451